Accugenix reviews identification methods for environmental monitoring in non-sterile production facilities, such as pharmaceuticals or nutraceuticals.
For manufacturers of pharmaceuticals, dietary supplements or nutraceuticals, the presence of bacteria, filamentous fungi and yeasts are usually a cause for concern. However, a well-designed environmental monitoring (EM) programme should detect the presence of these micro-organisms before product contamination occurs. When bacterial or fungal isolates are recovered from a production facility, it is important to be able to identify accurately the organism to the species, and possibly strain level, to track the potential origin of the contamination, mitigate it and avoid delays in product release or to complete investigations.
Accurate microbial identification requires significant and continuous process refinements, familiarity and expertise in interpreting data, consistent qualified methods of analysis and timely updates to organism libraries. Regardless of the method used, this information is only as reliable, or accurate, as the reference libraries used. To identify correctly a large percentage of the unknown isolates in EM programmes, the libraries must contain all species relevant to the manufacturing environment involved.1
Additionally, it is critical to update libraries continually and incorporate new entries to stay current with an ever-evolving microbial world, where taxonomic changes and novel species are described daily.
According to FDA2 and FAO3 guidelines, microbial monitoring of critical components, areas and personnel should include routine identification of isolates to the species level. Broadly speaking, three methods are currently used in commercial settings: genotypic, proteotypic and phenotypic (see Figure 1).
The FDA states that: “Genotypic methods have been shown to be more accurate and precise than traditional biochemical and phenotypic techniques. These methods are especially valuable for investigations into failures (e.g. sterility test; media fill contamination)”.4
Genotypic identification methods involve sequencing different regions of the micro-organism’s ribosomal RNA genes (16S, ITS or D2), resulting in identification to the species or occasionally the subspecies level.
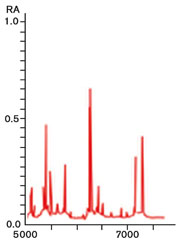
Figure 2: MALDI-TOF ribosomal protein spectrum
An emerging proteotypic technology in commercial bacterial identification for EM programmes is matrix-assisted laser desorption/ionisation-time of flight (or MALDI-TOF) mass spectrometry, which uses a ribosomal protein spectral fingerprint to identify bacteria to the species level.
Phenotypic technologies use either biochemical reactions or cell-surface component compositions to identify a micro-organism.
Each technology has advantages and disadvantages. The key is to balance the cost effectiveness of the technologies and to maintain a high level of accurate and reproducible identifications, while adhering to manufacturing guidelines and maximising the control over a production environment.
For identification of unknown microbial organisms, DNA sequencing rapidly provides data that are more accurate, robust and reproducible than relying solely on visual phenotypic characteristics. This is because the sequence-based result is not dependent on age and health of an organism, growth conditions or ancillary testing. In fact, samples can be viable or nonviable cultures or simply genomic DNA material collected from the microbe.
Genotypic identification
Genotypic methods use comparative sequencing of the ribosomal RNA (rRNA) region. The use of ribosomal DNA sequences for microbial taxonomic classification has been in practice for decades because the technology is inherently stable and thus allows for reproducible data for classification as well as for identification.
In all living things, the ribosome (the organelle that is the site of protein synthesis in the cell) contains different sized rRNAs, which are transcribed from ribosomal operons in the organism’s genome. The rRNAs fold into elaborate three-dimensional structures and are incorporated into an intricate protein-RNA complex that is critical for the ribosomal function and cell survival (see Figure 4).
Bacterial isolates are identified using the 16S rDNA region, because it is universally distributed among bacteria and contains species-specific variable regions. The identification of fungi, especially filamentous fungi, has historically been a very difficult task. Due to the amount of experience and time required to identify accurately filamentous fungi to the species level, it has been common practice to settle with either identifying these organisms to the genus level, or in some cases, simply identify them as “moulds”. Genetic approaches to fungal identification provide a more acceptable, reliable and rapid method for identification to the species level.
The internal transcribed spacer (ITS2) region is the ribosomal DNA region that is sequenced by fungal taxonomists, because the ITS2 region has a higher degree of variation between closely related species than other rDNA regions in fungi, such as the D2 region of the large subunit of the ribosome.5
To perform bacterial or fungal genotypic identifications, the target regions of the rRNA genes are amplified by PCR and sequenced (Figure 3). The sequence data are then analysed and aligned in order of increasing genetic distance to relevant sequences against a library database to achieve an identity match. Since interpretation of the data and the database against which the sequence is compared are both essential parts of the identification process, it is important not to neglect the method of data analysis and library coverage when choosing an identification system or provider.
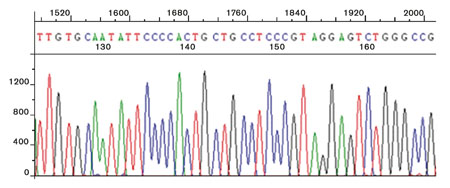
Figure 3: DNA sequencing chromatogram
Differences between systems include fully automated or manual interpretation of the data. Manual data analysis and alignment are extremely repeatable and result in the ability to correct for standard sequencing anomalies and sequence variations that can cause the data to appear to be mixed or of poor quality. Poor quality data are usually truncated in fully automated analysis programmes, which can lead to faulty interpretation of the data and incorrect identification of EM isolates or of bioburden samples taken from final product.
Proteotypic identification
In proteomics, a field devoted to studying the full set of proteins encoded by the genome, “proteotypic” describes a peptide sequence that serves to identify a particular protein. In microbial identification, “proteotypic” is used to indicate a protein spectrum that serves to identify a particular micro-organism. For industries that are required routinely to identify micro-organisms, the ideal technology is one that is accurate, reproducible, fast and inexpensive.
MALDI-TOF mass spectrometry exhibits all these traits. This proteotypic method has demonstrated increased accuracy and reproducibility over phenotypic systems.6 The analysis of a bacterial sample results in a unique protein spectral fingerprint that is then compared with a validated database for identification. The spectrum is composed of the ribosomal proteins of the bacteria, which are translated from DNA but are not subject to the expression variability seen in phenotypic methods. These ribosomal protein molecules are constitutively expressed and structural, so they are consistently present in the cell in very high levels.
MALDI-TOF analyses a small amount of bacterial sample from a fresh culture that is suspended in a solution containing a matrix and then dried to a target plate. The plate is placed in the vacuum source chamber and irradiated with a pulsed laser beam. The ionisation process utilises the matrix to absorb laser energy while protecting the proteins and transferring ions to the intact molecules. The proteins are released from the matrix and enter the gaseous state as charged molecules. The ions, accelerated by electric fields, are then separated based on the time it takes them to travel a specified distance – time of flight (TOF).
The lower the mass of the ion, the faster it will reach the detector. The protein spectrum or fingerprint that emerges from this process is then compared with a library of spectra from known bacteria. For effective identification, the library needs to contain relevant organisms found in manufacturing environments and must be continually updated to include novel organisms and taxonomic changes.
For situations where full sequence data are not required, such as for routine monitoring, but when a repeatable, accurate, cost-effective identification is still required, the MALDI-TOF technology is a good alternative when supported by a robust, relevant library for EM programmes. However, because the technology is in its infancy for identifications of micro-organisms found in a production environment with respect to library database development, a polyphasic approach using the proven method of rDNA sequencing is recommended when an identification cannot be made by MALDI-TOF.
Phenotypic identification
There are multiple systems that use phenotypic characterisation to obtain a microbial identification. These systems have been in use for decades and differentiate between organisms based on the results of biochemical tests, such as sugar fermentation, or physiological properties, such as salt or pH tolerance. One phenotypic method provides microbial identification based on cellular fatty acids that are extracted and methylated. The resultant methyl esters are separated by gas chromatography and their patterns are compared against a database.
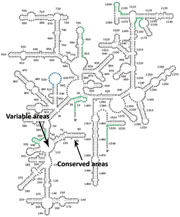
Figure 4: 16S ribosomal RNA structure for Escherichia coli
All of these phenotypic systems require a healthy organism, and depending on the technology used, may or may not need to be cultured on specific media and require ancillary testing, such as Gram stain, to achieve identification. The systems tend to be easy to use and have high throughput and have been, and continue to be, primarily used in clinical settings. However, micro-organisms isolated from manufacturing environments on compendial media will probably be physiologically stressed and may not fully express their phenotypic or biochemical characteristics, resulting in erroneous identification.
For many organisms, traditional phenotypic identification is problematic. Identification can be dependent on media and temperature used to grow the organism and can lead to subjective interpretation of the test results and a higher rate of inaccurate identifications. Not all strains within a given species consistently exhibit a particular characteristic, thereby limiting phenotypic identification methods. Additionally, library databases used in support of phenotypic identification are often limited and geared towards clinical isolates.
Despite their major shortcomings, these methods still have a role in a microbial quality programme. The results from the phenotypic reactions can help determine the biochemical activity of an organism on the product and the resulting stability of the product in the presence of that organism. Understanding the nutrient requirements of an organism can provide insight into controlling or eliminating the organism in the environment or preserving the product.
In conclusion, current available methods of identification range from genotypic and proteotypic to phenotypic, with 16S and ITS2 sequencing being the gold standards for bacterial and fungal identification, respectively, especially when combined with reference quality interpretation methods and curated libraries focused on organisms relevant to the dietary supplements, personal care products and other manufacturing industries.
The introduction of the proteotypic MALDI-TOF-based method of identification provides an additional highly accurate, fast and inexpensive option for routine monitoring programmes. When identifications are based on phenotypic characteristics, such as with biochemical analysis, the methods are more error prone, variable and subjective.
An EM programme should detect micro-organisms in a reproducible way, so as to monitor the state of control in the environment effectively. Consistent methods will yield an identification history that allows for comprehensive data comparison and interpretations. DNA sequencing provides the most consistent and unambiguous data set that is reproducible from lab to lab and over time. Genotypic and proteotypic identification are unlike phenotypic, which can be affected by differential gene expression resulting in variable characteristics. Additionally, the DNA sequence is stable and unchanging and is a tool for identification as well as a unique descriptor for the micro-organism that can be used for tracking and trending.
References
1. Bacterial Library Listing and Comparison. White Paper, Accugenix (2011).
2. FDA Regulation 21 CFR Part 111 – Current Good Manufacturing Practice in Manufacturing, Packaging, Labeling, or Holding Operations for Dietary Supplements (2011).
3. FAO/WHO Joint Working Group Meeting, Guidelines for the Evaluation of Probiotics in Food (2002).
4. FDA Regulation 21 CFR Part 211 – Current Good Manufacturing Practice for Finished Pharmaceuticals (2011).
5. ITS DNA Sequencing for Fungal Identification. White Paper, Accugenix (2010).
6. The AccuPRO-ID Solution from Accugenix, Inc. White Paper, Accugenix (2011).




