Although the majority of environmental isolates we receive from pharmaceutical manufacturers are bacterial, fungal contamination is a significant concern for the sector. Fungi can cause spoilage of products and harm to patients. They can enter facilities in several ways – airborne spores can enter through the smallest of openings, or fungi can be carried into facilities with raw materials or by employees.
Once they have made their way in, they are ideally equipped to exploit any damaged and porous surfaces or damp cardboard packaging. Consequently, controlling and monitoring fungi in cleanroom environments is an important activity. However, fungal identification can be more challenging than identification of bacterial isolates, and this can potentially make the task more difficult.
Fungal identification can be more challenging than identification of bacterial isolates
So what are the most identified fungi from pharmaceutical manufacturing and cleanroom environments? Over the years we have identified thousands of unknown fungal isolates for our customers, and collating the results may give a valuable insight into what we have found to be the most commonly isolated genera and species.
It should be noted that the data presented in this article do not represent a controlled scientific study, and we hold no information on the type or grade of the facility or the area within which the isolates were obtained. We have however collated the results of almost 4000 fungal identification results made over 15 years. The majority of these have come from pharmaceutical manufacturing environments, but the data also includes results from some other sources including food and drink, cosmetic/personal care product manufacturers and some other sources.
The fungi that we have identified come from 56 different genera but our “top 20” most common identification results represent more than 90% of the total identifications, and Figure 1 focuses on these. Looking at the chart, three genera stand out – together accounting for more than half (63%) of all identifications – these are Cladosporium, Penicillium and Aspergillus.
Another key point highlighted by the chart is the high proportion of isolates that have returned an identification result that lists more than one genera – most notably Cladosporium/Mycosphaerella. I will return to this point later, but looking at the bigger picture, the top three genera are all known to be ubiquitous airborne moulds, and together include hundreds of different species, so it is no surprise that they are the most commonly identified isolates.
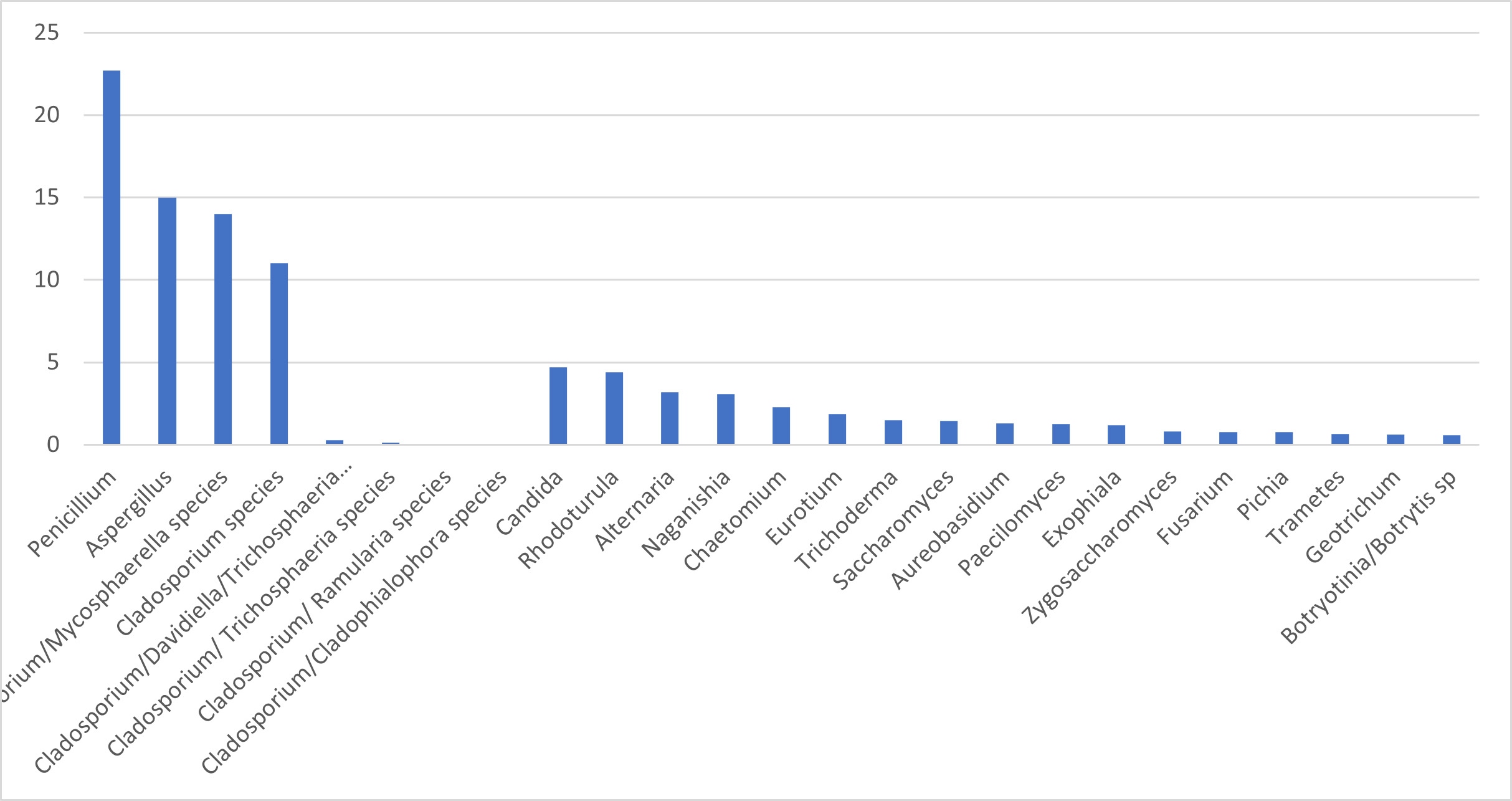
Figure 1
Although our data is not obtained exclusively from cleanrooms, our most commonly identified isolates tie in well with a recent paper that presents a study of fungi isolated from Grade C and D cleanrooms over a ten-year period (Sandle, 2021). The isolates in that study were drawn from an environmental monitoring regime designed to recover both bacteria and fungi. One hundred and eighty fungal identifications were undertaken, and of these, 133 were able to be characterised to either genus or species level using visual and phenotypic methods.
The most common identifications in this study also included Penicillium and Cladosporium, however while Sandle (2021) found that Fusarium was the third most commonly identified isolate, at 18%, Fusarium species have accounted for less than one percent of the fungal isolates identified by NCIMB. Aspergillus was a significant genus in both data sets – representing 15% of NCIMB identification and 8% in the Sandle (2021) study.
All five of the genera listed in Table 1 are very commonly found in the environment, and, in addition to the fact NCIMB’s identifications are not restricted to cleanroom isolates, some variation in their relative proportions might be expected due to variations in the local environment and in a smaller data set, one atypical incident might result in a higher proportion of a particular genus.
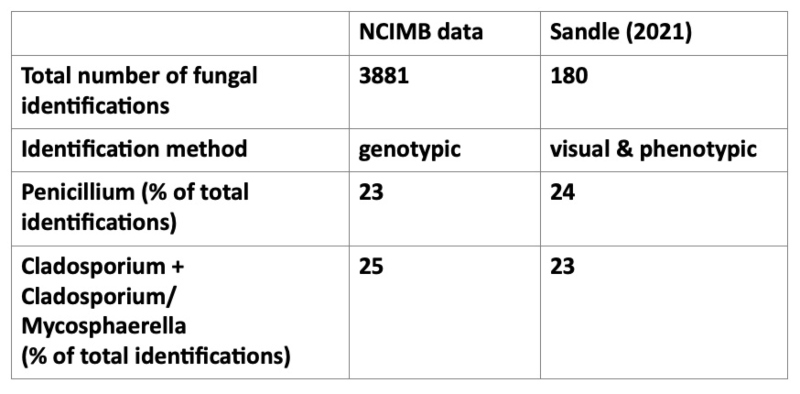
Table 1
While Sandle (2021) used visual and phenotypic methods for identification, a genotypic method was used at NCIMB, and we can look at the breakdown of identifications within each genus.
Species vs genus level identification: Penicillium
Looking more closely at the identification results highlights that for Penicillium, the majority of identifications returned a genus rather than a species level match, and this is illustrated in Figure 2. Twenty-nine individual species were identified – the top ten represented 96% of all Penicillium identifications and these are listed in Figure 2.
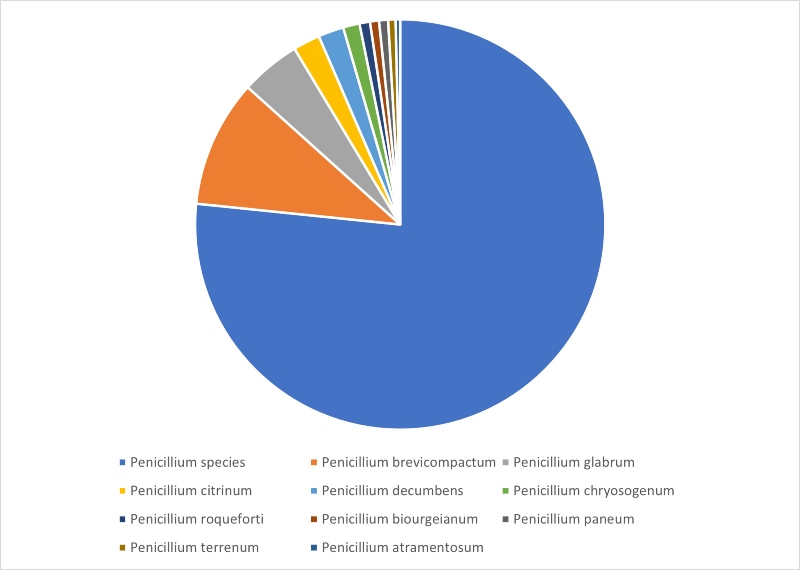
Figure 2
In many cases I find that the section of DNA sequenced to make the identification – the D2 region of large-subunit ribosomal RNS gene sequences (D2LSU) - gives a 100% match to more than one species. For example, a single isolate sequence often matches at 100% to Penicillium camemberti, Penicillium clavigerum, Penicillium commune, Penicillium corylophilum and Penicillium crustosum. D2 is not the only sequence used for genotypic fungal identification, but it is our first port of call because the validated MicroSEQ database can be used to make identifications.
Another potential route is to sequence a different region, and the internal transcribed spacer is commonly used for this purpose. I have found this method sometimes returns a species level match when D2 sequencing has given a genus match - in one example the isolate was identified as Penicillium crustosum with a 100 % match. The database used for identifying isolates using ITS sequence data was more extensive than the MicroSEQ D2 database, but unvalidated.
Species vs genus level identification: Cladosporium
Looking in more detail at Cladosporium identifications shows that species level identification is not always straightforward for this very commonly isolated genus either. In fact, in many cases D2 sequencing alone cannot distinguish between Cladosporium and different, but closely related, genera. This is illustrated in Figure 3, which shows that more than 50% of identifications matched to more than one genus, with a further 42% matching to several different Cladosporium species.
It is quite common for a single isolate to match to Cladosporium asperulatum, Cladosporium chalastosporoides, Cladosporium cladosporioides, Cladosporium delicatulum, Cladosporium exasperatum, Cladosporium flabelliforme, Cladosporium funiculosum, Cladosporium gamsianum and Cladosporium globisporum at 100% similarity when using the MicroSEQ D2 database. These are the top ten matches listed by the software we use but it is possible the data may also match at 100% to further species.
Where an isolate matches to both Cladosporium and Mycosphaerella, I often find that it matches to Cladosporium cladosporioides, Cladosporium herbarum and Mycosphaerella aronici.
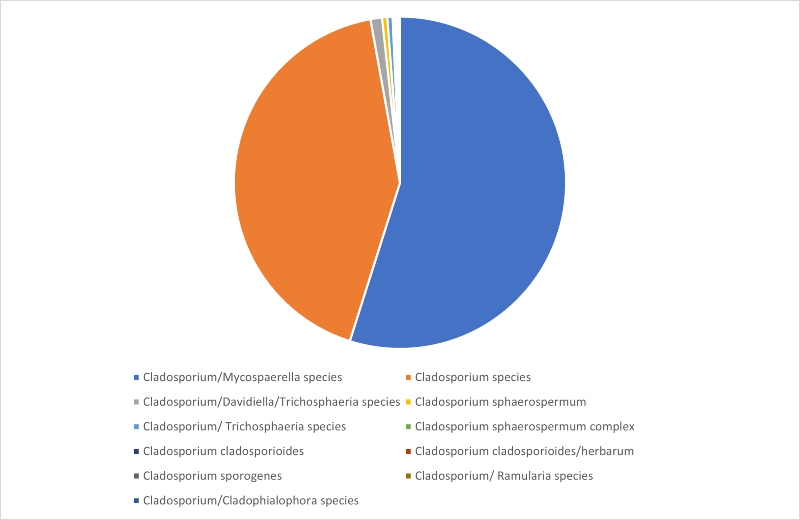
Figure 3
Again, ITS sequencing may return a species level match, for example, one isolate which matched to Cladosporium cladosporioides, Cladosporium herbarum, Cladosporium oxysporum, Mycosphaerella aronici and Mycosphaerella tassiana, was identified as Cladosporium cladosporioides using ITS sequencing together with an extensive, but unvalidated database of published sequences.
Species vs genus level identification: Aspergillus
Looking at our third most commonly isolated genus, Aspergillus, Figure 4 shows that, in contrast to the results seen for Cladosporium or Penicillium, D2 sequencing returned a species level identification for 75% of identifications. The most commonly identified Aspergillus species was Aspergillus versicolor, followed by Aspergillus fumigatus and Aspergillus sydowii. Aspergillus species are found in soil, can cause food spoilage, and are also commonly found in damp indoor environments and all three of these species have been associated with disease in humans.
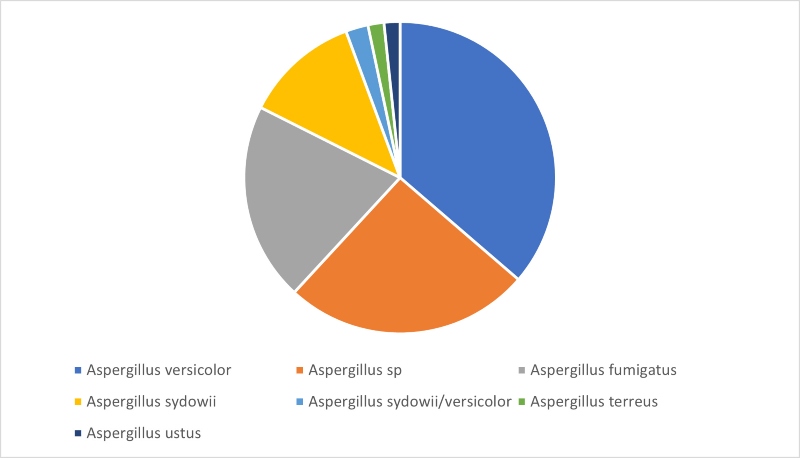
Figure 4
Conclusions
Reviewing historical data illustrates that the most commonly identified fungal isolates are broadly in line with what might be predicted from the prevalence of those genera in the wider environment, and the ease with which spores disperse. The results are also broadly in line with a previously published study designed to assess the most common fungal isolates from cleanrooms.
The data highlights that the most common genera isolated can be quite difficult to identify at the species level, using D2 sequencing. However, In many cases a genus level identification may be sufficient. This is a decision that has to be made in each individual case - the question to ask is perhaps will the action taken differ if a species level match is obtained. If not, then a genus level identification may be sufficient. But if a species level identification is required, perhaps to investigate a particular issue, we have found sequencing a different section of DNA can deliver a species level result.
